AWARDEES: Barnett Rosenberg, Loretta VanCamp, Thomas Krigas
FEDERAL FUNDING AGENCIES: National Institutes of Health, National Science Foundation
Barnett “Barney” Rosenberg wasn’t a cancer researcher — but he and his lab team helped unlock a breakthrough cancer treatment. In the 1960s, Rosenberg, alongside lab technician Loretta VanCamp and a team of graduate students including Thomas Krigas and a few postdocs, was examining how electric fields affect cell division in E. coli bacteria. To their surprise, the bacteria stopped dividing and instead elongated into spaghetti-like shapes — a striking, unexplained phenomenon they investigated further. After a couple years of follow-up experiments, they discovered the true cause: Platinum compounds released from the electrodes, not the electric field itself. This serendipitous finding led to the development of cisplatin, a platinum-based chemotherapy drug approved in 1978. At the time, the idea of using a metal-containing compound in medicine was unconventional and met with skepticism due to concerns over toxicity to humans. After harmful side effects were mitigated, cisplatin was approved and delivered unprecedented results — most notably, increasing the survival rate for testicular cancer from around 10% to over 90%. Its success transformed cancer treatment and has saved countless lives.
Barnett Rosenberg
Barnett Rosenberg, A Creative Mind
After serving as a pharmacist in the Philippine Islands during World War II, Barnett Rosenberg returned home and earned his Ph.D. in Physics from New York University in 1955. Rosenberg was a deeply curious and imaginative thinker, known for spending hours listening to classical music while contemplating scientific problems. His daughter, Tina Rosenberg Varenik, recalled “sitting and thinking made him more creative.” Rosenberg even had a connection to Albert Einstein, who advised Rosenberg’s Ph.D. advisor, which made Rosenberg an “academic grandson” of the “Ultimate Thinker.”
A core tenet of Rosenberg’s research philosophy was his dedication to exploring science with the goal of advancing human well-being. In 1961, he joined Michigan State University, where he co-founded the biophysics department. His work focused on the fundamental biophysical mechanisms that govern cellular behavior — how cells respond to stimuli, divide, and interact with external forces. Driven by his curiosity, Rosenberg approached biological questions through the lens of physics, laying the groundwork for a uniquely interdisciplinary and unconventional research style. This perspective enabled him to uncover insights that more traditional approaches might have overlooked. Although Rosenberg was not formally trained in cancer research, his unique viewpoint proved to be a powerful asset when his lab would ultimately synthesize two of the three FDA-approved platinum-based chemotherapy drugs still in use today.
Rosenberg’s Lab and a Not So “Electric” Discovery
At Michigan State University, Rosenberg’s early research was supported by a National Science Foundation grant and focused on understanding the electrical properties of proteins, as well as vision-related studies involving rhodopsin — a light-sensitive protein in the retina — and carotenoids, naturally occurring pigments. The team of mostly postdoctoral students in Rosenberg’s lab took delight in the carotenoid work, which involved extracting the pigments from the shells of boiled lobsters. Since the lobster meat had no scientific use, it was happily enjoyed by the lab staff.
Loretta VanCamp and Barnett Rosenberg
Early in his tenure, Rosenberg hired Loretta VanCamp to be his lab director. She was regarded as intelligent and perceptive, and if not for the gender-based barriers in academia at the time, may have well been a doctor. VanCamp had remarked, “I chose med tech when they told me that was as close to becoming a doctor as I could without going through the hassle of a woman trying to become a doctor in a man’s world.”
In 1963, VanCamp initiated a research project to investigate the effects of electric fields on dividing E. coli bacteria. While the idea originated with Rosenberg, it was VanCamp who carried out all the experimental work — a testament to her role in the lab but also a reflection of Rosenberg, who avidly promoted the work of other young scientists and women. The experiment sought to address a speculative notion held at the time that a magnetic component might be operating in mitosis (a type of cell division). Shortly after the electric field was generated, a project-altering and unexpected observation was made — the single-celled bacteria had undergone filamentous growth. This observation was understood immediately to mean that cell division had stopped, but cell growth had not. The “spaghetti-like shaped” cells reached up to 300 times their normal length. While this result was surprising, there was still much to learn about what caused it, and an intensive search began. Most thought this work could help model and prevent bacteria growth, but Rosenberg believed if they could find out what was causing this response in the bacteria cells, it may translate to a useful anticancer agent in human cancer cells.
Loretta VanCamp and Barnett Rosenberg
By the fall of 1964, Rosenberg’s lab was continuing experiments to identify what had caused the filamentous growth the year prior and evaluating the role of many chemical compounds as possible causative agents. As part of the continued quest, Thomas Krigas, a chemistry graduate student working part-time in the lab, relayed to Rosenberg a hypothesis that the platinum molecules present in the culture medium of the E. coli bacteria were undergoing electrolysis due to the presence of the electrical field. The lab’s work shifted to focus on looking for chemical compounds that might be produced due to the chemical reaction. Over the next several months, Krigas would help lead the way to isolate and identify the active chemical agent affecting cell division, a platinum electrode compound known as cis-diamminedichlorideplatinum [Pt(NH₃)₂Cl₂], later named cisplatin. While Krigas helped synthesize the compound, VanCamp observed that the filamentous growth resulted not from the electric field itself, but because of the products formed during electrolysis at the platinum electrodes. Rosenberg, VanCamp, and Krigas published a Nature article on the “Inhibition of Cell Division in Escherichia coli by Electrolysis Products from a Platinum Electrode” in February 1965 and were all credited with the discovery. The identification of the compound laid the groundwork for subsequent development and testing.
Cancer Research and the Development of Cisplatin
At the time of the early work on cisplatin in the 1960s, much about cancer — its origins, and potential treatments — remained unknown. The first chemotherapeutic agents emerged from research on chemical warfare and mustard gas during WWII. Chemically based therapeutic drugs, known as chemotherapy, had shown early success in addressing childhood leukemia and Hodgkin’s disease. However, skepticism about their overall effectiveness persisted as cisplatin entered the scene.
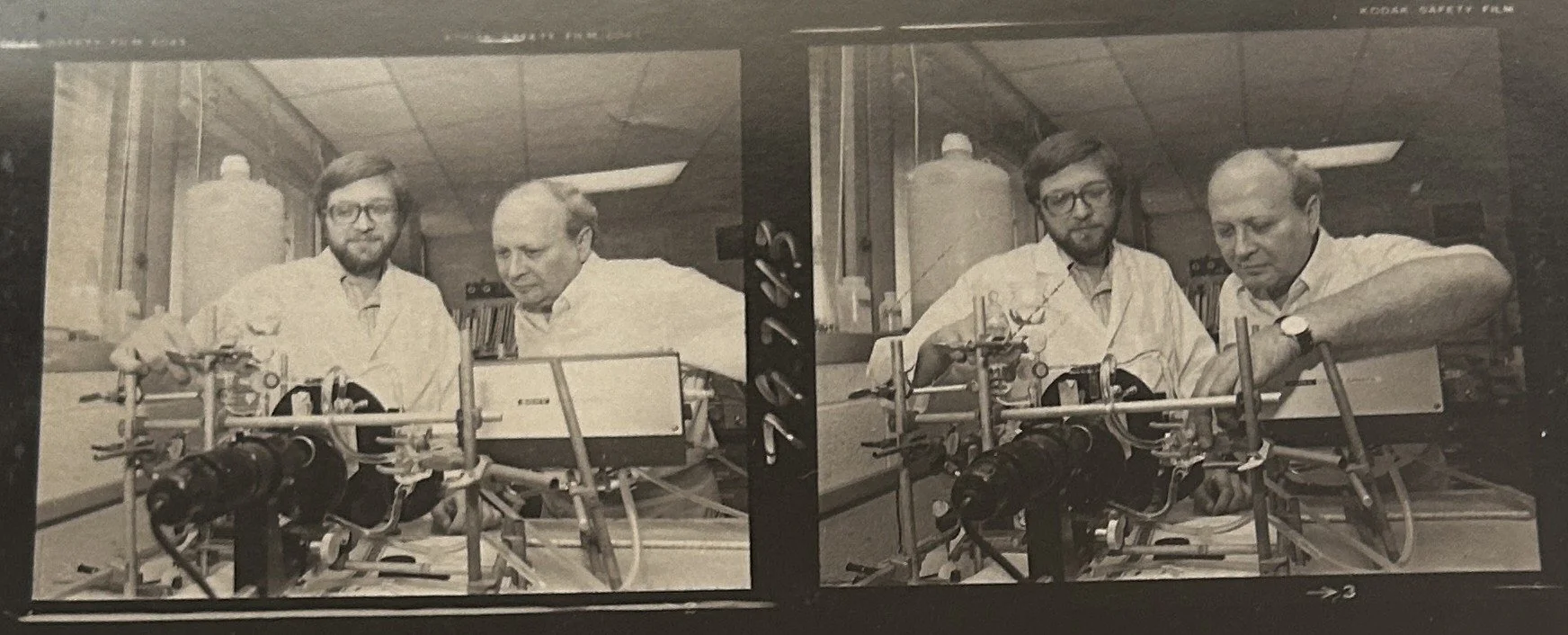
Dr. Rosenberg and graduate student in lab
Following the publication of the 1965 Nature paper, Rosenberg’s lab began testing cisplatin in mice. Although the team hoped their initial findings would generate widespread interest in the research community, it didn’t quite pan out that way. In 1968, Rosenberg made what he later called a “purely intuitive jump” by testing cisplatin on mice bearing solid tumors, known as Sarcoma 180 (S180). Despite its high toxicity — particularly kidney damage at elevated doses — the mice tolerated low doses of cisplatin, and their tumors shrank. Six months later, the mice remained healthy, with no tumor recurrence. In effect, they were “cured.” In 1969, Rosenberg and VanCamp, along with collaborators James Trosko and Virginia Mansour, published a second Nature article, “Platinum Compounds: A New Class of Potent Antitumor Agents,” detailing these promising results. This time, their findings would gain more attention.
The Fight for Clinical Trials
After observing positive responses to cisplatin in mice, interest in Rosenberg’s work grew, particularly among international researchers. As he shifted his focus to advocating for clinical trials, skepticism persisted within the medical community. The idea of using a metal-containing compound in medicine, coupled with concerns about the drug’s toxicity, led some to question cisplatin’s viability as a cancer treatment. Both the FDA and the National Cancer Institute (NCI) needed to be convinced that the drug’s risk-benefit ratio was acceptable. While early clinical trials in the U.S. began in 1972 with funding from the NCI, the first Phase 1 clinical trial of cisplatin took place in England in 1971. By 1973, early clinical findings were ready for publication and were presented at the first open international platinum meeting held at Wadham College in Oxford. Some studies reported encouraging results, including a study of 20 women in England with advanced ovarian cancer, where cisplatin successfully shrank their tumors. Another study in New York involving 11 patients with testicular cancer — who had previously been treated with other drugs — showed partial response to cisplatin, with three patients achieved complete remission.
However, these promising outcomes were overshadowed by serious concerns about kidney toxicity, which posed a potentially significant limitation. The toxicity issue nearly halted cisplatin testing altogether until further research could be done to address these risks. Physicians pioneered the “mannitol diuresis procedure,” learning that infusing patients with large volumes of fluid during drug administration could effectively “wash out” the kidneys and protect them from damage. This approach proved successful and was quickly adopted by other clinicians. By 1975, cisplatin entered Phase II clinical trials, following a reduction in kidney-related concerns.
Cisplatin was ultimately approved by the FDA in 1978 for treatment of ovarian and testicular cancers. The development of the mannitol diuresis procedure helped mitigate drug-related kidney toxicity and enabled broader, safer use of cisplatin. Despite FDA approval, pharmaceutical companies remained reluctant to license or market the drug, even though all initial clinical studies had been underwritten by NCI. Rosenberg continued to face resistance and spent years persuading medical professionals that toxic platinum could effectively combat tumors without endangering patients.
A Lasting Legacy
Before the introduction of cisplatin, approximately 95% of young men diagnosed with testicular cancer in the 1970s died from the disease. At the time, testicular cancer was considered a death sentence. Today, however, nearly every patient can be successfully treated. One notable case is that of John Cleland, the first patient “cured” of metastatic testicular cancer. Mere days from death, his doctor decided to add the experimental drug — cisplatin — to a cocktail of chemotherapy drugs. Cleland was 23 years old when he was diagnosed in November 1973, but went on to live a normal life, passing away in 2022 from unrelated causes at the age of 71. In 1999, the results of three clinical studies showed that cisplatin, when combined with radiation, reduced death rates from cervical cancer by up to 50%. These findings, published in The New England Journal of Medicine, were so compelling that the NCI contacted oncologists worldwide before the official publication, urging them to adopt the treatment. Cisplatin has been found most effective in regiments treating testicular, ovarian, bladder, cervical, head and neck, and lung cancers.
Article in local newspaper reporting on Dr. Rosenberg’s death in 2009
Beyond FDA approval, research has continued to improve patient tolerance of cisplatin and mitigate its side effects. Additional work in Rosenberg’s lab led to the synthesis of carboplatin, a milder platinum-based drug that reduces side effects, particularly in patients with pre-existing kidney or hearing issues. To date, two of the three FDA-approved platinum drugs were synthesized in Rosenberg’s lab. Cisplatin remains a cornerstone of chemotherapy protocols. Researchers are still developing new analogs to reduce toxicity, tailor treatments to specific cancer types, and combat drug resistance. Once destined for the pharmacologic graveyard, cisplatin is now used in the initial treatment for several types of cancer and has been listed on the World Health Organization’s List of Essential Medicines since 1984. Its discovery also spurred advancements in other areas, including development of anti-nausea medications, studies of DNA repair mechanisms in cancer cells, and broader research into metal-based drugs.
For Rosenberg, VanCamp, and Krigas, their work on cisplatin defined their careers and earned them numerous recognitions. Many believed Rosenberg was destined to receive the Nobel Prize; although he was nominated several times, he never received the “ultimate prize.” He passed away in 2009 at the age of 82, leaving behind a body of work that has saved countless lives. Before her death in 2006, VanCamp was honored with an honorary Doctor of Science from Michigan State University in 1999. In 2019, the MSU Research Foundation—funded in part by royalties from the cisplatin and later the carboplatin discoveries—opened the VanCamp Incubator and Research Labs to support emerging companies in the Lansing, Michigan area focused on life sciences, biotechnology, and physical sciences. Krigas passed away in 2023. His obituary reads, “As Tom would say, it was just dumb luck that the team's efforts would result in the development of the first major cancer chemotherapy drug, Cisplatin.” What may have begun as a stroke of “dumb luck” became a profound breakthrough in cancer treatment, and proof that curiosity, collaboration, and perseverance can change the course of medicine and save millions of lives.
By Meredith Asbury